P{UAS-RGB.cct} has the following structure.
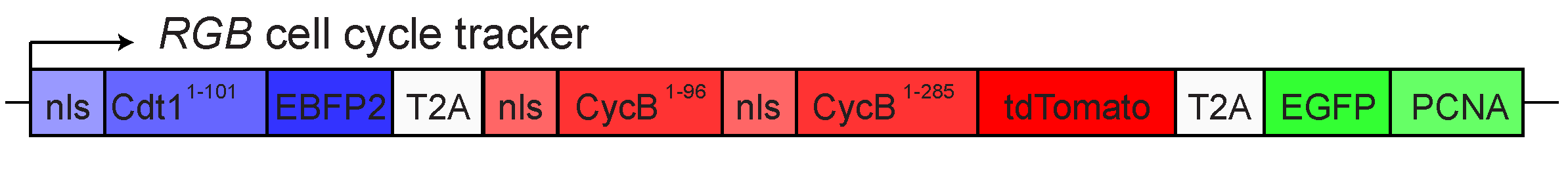
As shown in the figure below, it expresses:
• EBFP2 with a nuclear localization signal fused to a degron sequence from dup/Cdt1 for degradation during S phase.
• tdTomato nuclear localization signals fused to degron sequences from CycB for degradation during late mitosis and early G1 phase.
• EGFP fused to PCNA to give a unique subnuclear localization during S phase.

It was described in Handke et al. (2014).” Towards long term cultivation of Drosophila wing imaginal discs in vitro” PLoS ONE 9: e107333.
P{UAS-RB.cct} [No FBid yet] is identical except for omission of the T2A-EGFP-PCNA sequence.
These constructs provide alternatives to the fluorescent ubiquitination-based cell cycle indicator (Fly-FUCCI) for marking cell cycle stages.

