The Super Recombinator Cas9 with Serine Recombinase (SuRe-CR) system was designed by Junjie Luo and Mark Schnitzer to recombine insertions in the same attP landing site onto the same chromosome with simple genetic crosses utilizing one of three serine recombinases (phiC31, Bxb1 or TP901-1). Two to many insertions can be put in the same landing site with successive rounds of recombination.
Briefly, Recombinator 1 (R1) and Recombinator 2 (R2) strains carrying Cas9 are used to put adaptors next to transgenes of interest in attP landing sites. R1 strains add Adaptor 1 (AD1) between a transgene and the attL. R2 strains add Adaptor 2 (AD2) between a transgene and attR. A serine recombinase (phiC31, Bxb1 or TP901-1) is then used to recombine the two together via the attP and attB sites provided by AD1 and AD2, respectively. See Luo et al 2025 for details.
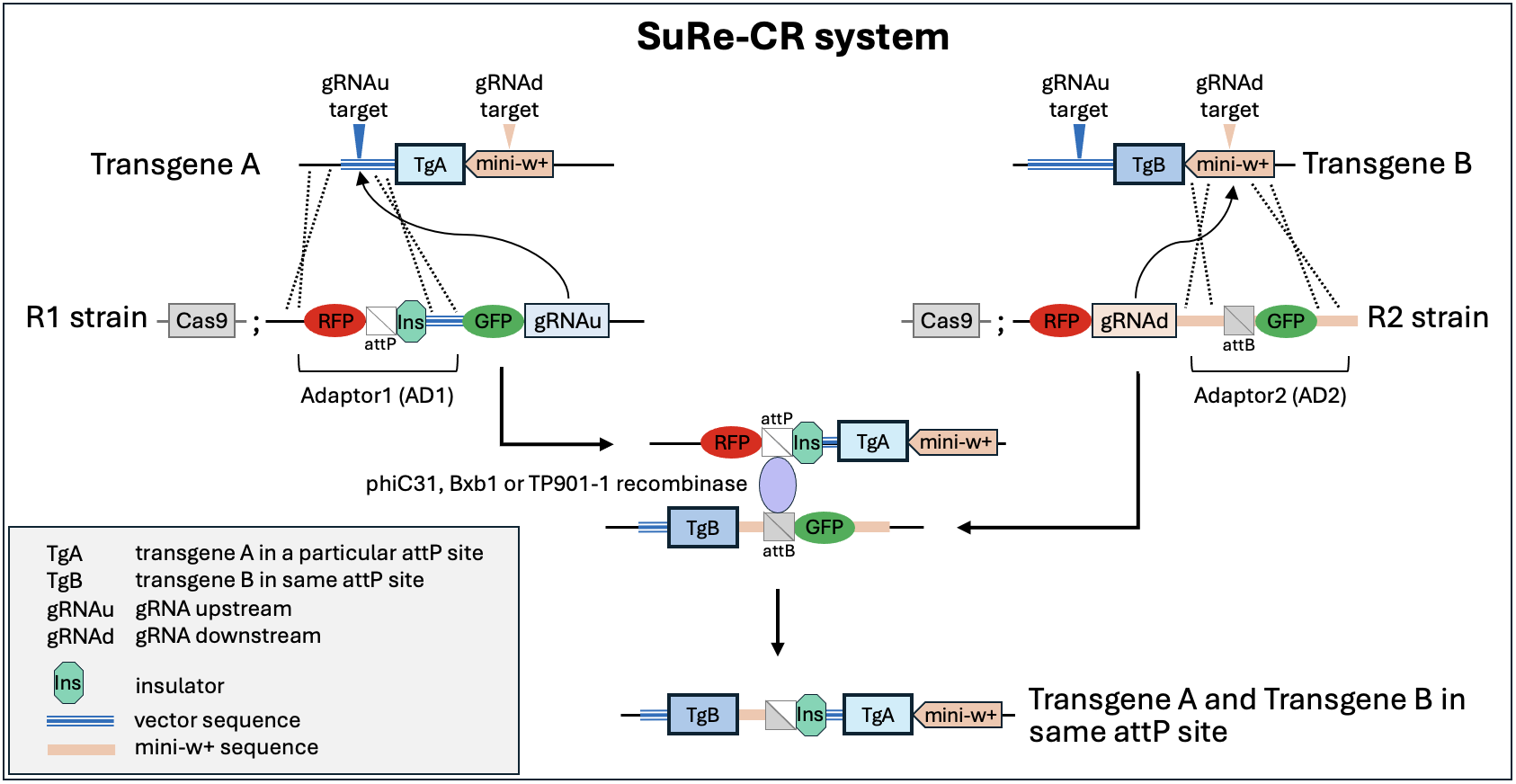
R1 and R2 strains use gRNAs and homology regions to target adaptors to sequences flanking transgenes. Different transgenes have different backbones so be sure to pick the right R1 and R2 strains for your particular transgene and landing site. For a brief primer on what R1 and R2 target, see this page.
We have information on the adaptors donated to the BDSC in a downloadable 'sure_adaptors' file in Excel or csv form. Information includes the sequences of the gRNAs, what markers are present, some information on the homology regions present, and, for those R strains that target mini-w+, the fate of the mini-w+ after AD insertion.
R1 and R2 transgene sequences can be found on the SuRe sequences page.
Schnitzer lab has also created lines for HACKing an adaptor plus coding sequences for a split GAL4 component into an existing GAL4 insertion in one step. See the SuRe-HACK page for more information.

