Genetic underdominance at a locus occurs when heterozygotes have lower fitness than both homozygotes. Underdominance is interesting with respect to disease vector control strategies, because underdominant genetic elements cannot become fixed in a population unless they are introduced at frequencies above a threshold. These population dynamics potentially allow a transgenic construct to be introduced into a population with confidence that it could never become permanently established—or if it were deliberately established that it could be deliberately eliminated.
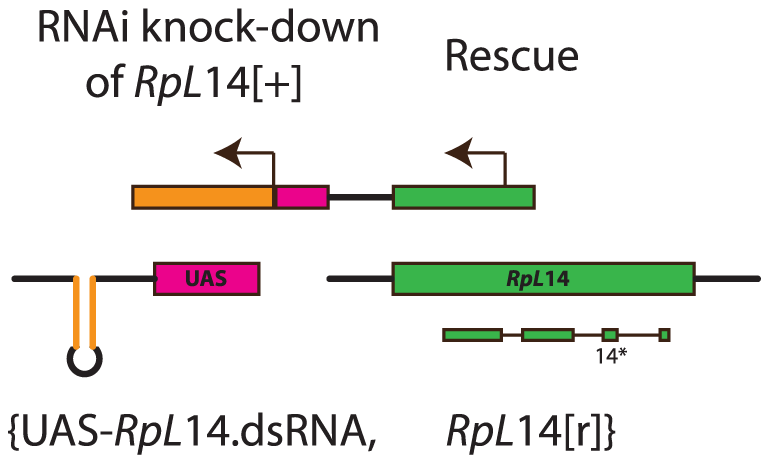
Reeves et al. (2014) generated the M{UAS-RpL14.dsRNA.RpL14[r]} construct that produces healthy homozygotes, but nearly sterile heterozygotes in the presence of ubiquitously expressed GAL4. Consequently, fully fertile individuals in a population are either homozygous for the construct or a wild type chromosome. As shown in the figure, the UAS construct expresses a dsRNA against the RpL14 gene. It also expresses, under its own regulatory sequences, a RpL14 gene modified to be insensitive to the dsRNA through the substitution of 14 nucleotides. Heterozygotes have low fertility because RNA interference eliminates RpL14 protein generated from the normal chromosomal locus, but the modified transgenic RpL14 gene is not expressed at high enough levels to allow more than marginal fertility. (Two copies of RpL14 are required for fertility; flies with a single copy typically show Minute phenotypes including sterility.)
The following stock were used by Reeves et al. (2014).

