SPARC (Sparse Predictive Activity through Recombinase Competition) is a method that uses phiC31 recombinase to stochastically catalyze irreversible recombination between two possible attP/attB combinations to limit expression levels of an effector (Isaacman-Beck et al., 2019). One attP/attB combination removes a STOP cassette, allowing expression of an effector (Rxn 1 in panel a). The other combination preserves the STOP cassette (Rxn 2 in panel a).
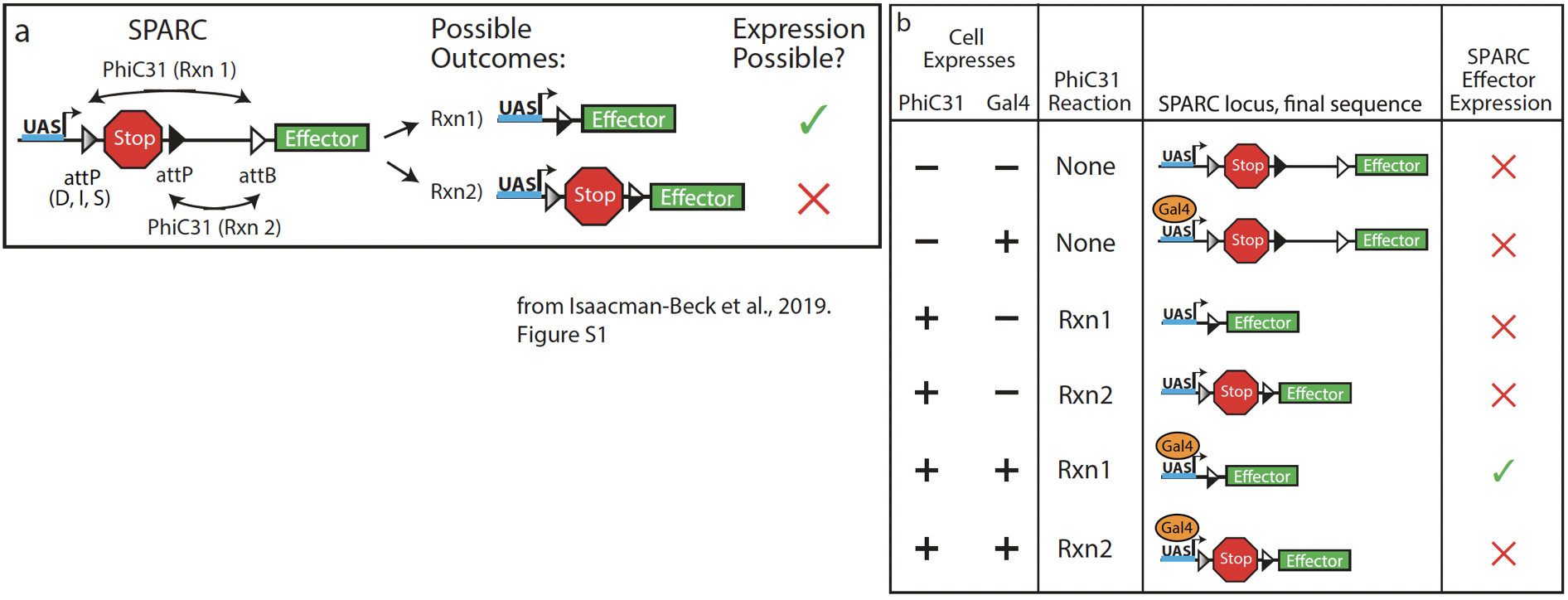
When the two attP sites are identical, these reactions happen at approximately equal probability, thereby producing ~45% sparsening. Truncating the attP site from its full length (attP60) to shorter sequences (attP38 and attP34) decreases the efficiency of the phiC31-mediated recombination. When attP38 or attP34 are used for Rxn 1 and are paired with full length attP60 for Rxn 2, the reaction between the attB and the full length attP60 is more likely. The construct with attP38 results in ~15% of cells expressing the effector, and the construct with attP34 results in ~5% of cells expressing the effector. They named these three modules SPARC-D, SPARC-I, and SPARC-S (D = “Dense”, I = “Intermediate”, and S = “Sparse”).
See the SPARC_user_guide for sample crossing schemes and more information on using these stocks.
With some SPARC-effector pairs (LexA.p65, mCD8::GFP), they saw leaky expression of the effector in the absence of phiC31 and generated a new version, called SPARC2, that includes ribozyme sequences. They find that these sequences decrease leaky expression up to ~10,000-fold and ensure that the transgenes express in the appropriate proportions of neurons.

