Zinc-finger nucleases create double-stranded DNA cuts at specific sites based on dimerization of the FokI endonuclease. Each nuclease monomer is localized via zinc-finger motifs binding to genomic sequences flanking the cleavage site as shown below for monomers directed to the yellow gene.
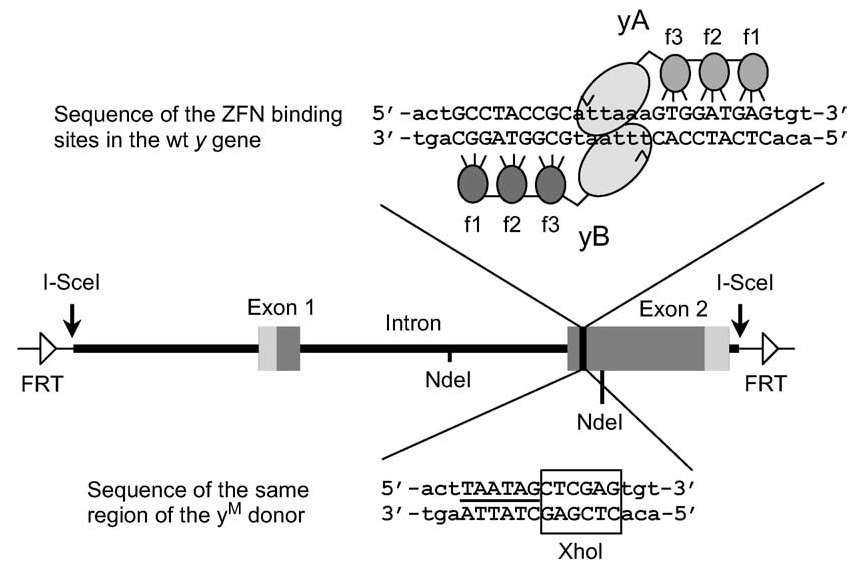
The figure also shows the structure of a repair template used to introduce a new sequence into the yellow gene. This template can be excised in vivo from the transgene P{y[M.donor]} by FLP recombinase and either used in a circular form or linearized by the I-SceI endonuclease.
Experiments involving cleavage of yellow and rosy by zinc-finger nucleases and their repair with template constructs are described in Beumer et al. (2006). "Efficient gene targeting in Drosophila with zinc-finger nucleases".
The following table shows stocks expressing zinc-finger nuclease monomers for cleavage of yellow. It also shows a stock with P{y[M.donor]}.

